Uni-Bio lab
The Uni-Bio Lab is a part of the Biocomplexity group, CMOL (Center for Models of Life) located at the Niels Bohr Institute in Copenhagen. We use mathematical models and quantitative experiments to distill simplicity behind biological complexity.
The Uni-Bio Lab is a part of the Biocomplexity group, CMOL (Center for Models of Life) located at the Niels Bohr Institute in Copenhagen.
Why?
We are interested in uncovering the unifying principles in complex biological systems. We like asking questions and look for simplest ways to answer them.
How?
We combine theoretical and computational models with quantitative experiments (primarily time-lapse). We enjoy interacting with our collaborators across physics, biology and medicine.
What?
Our current focus is on what can cells achieve collectively, how far can they go beyond what single cells can do on their own. A few examples are: ways to counteract stresses, balance their relative abundances in developing organs; encode and send the information across tissue and form complex and stable shapes in organs and organisms. More about this under Research Projects.
Life is a collection of processes on many different scales: milliseconds to centuries, nanometers to kilometers. As our capacity for information processing is finite we naturally zoom in and out between these scales to build a mental model of the world:
We pick up a few key elements and coarse-grain the details. In our group we use a similar approach of a coarse-grained modeling to capture complex biological phenomena.
The focus of our research is on identifying common principles in stress responses and development (in collaboration with StemPhys).
We find that the models at a right level of description reveal similarities across systems and organisms. These show up when we study:
- How receptor adaptation is encoded by networks that contain at least one of the two core circtuits
- How excitable dynamics can explain how the same proteins can encode opposing cell fates: life-death decisions and p53 dynamics in single cells; acute vs. chronic inflammation and IL-1/TNF/NF-kB dynamics in cell populations.
- How asymmetric damage segregation is a population strategy to rapidly adapt to stresses
- How right proportions are maintained during embryonic stem cell and pancreas differentiation
- How cellular polarities play together to form complex shapes (tubes and folds) in organs and organisms.
Stem cell differentiation
How do cells communicate and coordinate their fates to arrive at right proportions of differentiated cell types? How flexible/reversible are the transitions between differentiation states? Are there common principles (despite different set of proteins) at different differentiation stages and across organs?
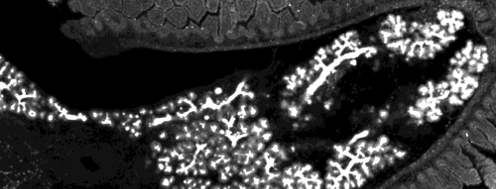
We address these questions in collaboration with Joshua Brickmann (ES cells, early blastocyst) and Anne Grapin-Botton (pancreas) labs theoretically by in-silico models and experimentally by quantitative single-cell imaging and rna and protein profiling.
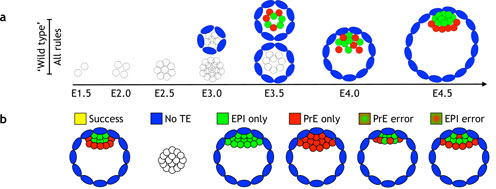
Stress responses
We strive to quantitatively understand the logic behind stress responses and how it unfolds itself in time and across cells in the tissue. What mechanisms did cells and organisms evolve to deal with the insults that perturb their normal function (stresses)?
What are the differences and similarities between the stress responses? (Both at the level of regulatory network and the dynamics of its components in single cells and on a cell-population level).With this knowledge and with the help of mathematical modeling the hope is to outline the "core circuits" that govern a given process. For this approach we employ a variety of theoretical tools complemented with experimental techniques (e.g. time-lapse of fluorescently tagged proteins in single cells) whenever possible.
Complex systems
We use agent based modeling to capture the possible mechanisms of species coexistence, epidemics and concurrent immunity spread of multiple competing diseases, or to explore the role of bacterial immunity (CRISPR) in phage-bacteria ecosystem. We have also developed a number of tools to analyze complex networks from an informaton perspecitve.
Species coexistence and bacterial immunity (CRISPR)
-
Targeted bacterial immunity buffres phage diversity. / Härter, Jan Olaf Mirko; Trusina, Ala; Sneppen, Kim.In: Journal of Virology, Vol. 85, No. 20, 21.08.2011, p. 10554-10560.
-
Ecosystems with Mutually Exclusive Interactions Self-Organize to a State of High Diversity. / Mathiesen, Joachim; Mitarai, Namiko; Sneppen, Kim; Trusina, Ala. In: Physical Review Letters, Vol. 107, No. 18, 25.10.2011, p. 188101.
Epidemics and concurrent immunity spread of multiple competing diseases
A minimal model for multiple epidemics and immunity spreading. / Sneppen, Kim; Trusina, Ala; Jensen, Mogens Høgh; Bornholdt, Stefan.In: PLoS ONE, Vol. 5, No. 10, 18.10.2010, p. e13326.
Information perspective in complex networks
-
Networks and Cities: An Information Perspective. / Rosvall, M; Trusina, Ala; Minnhagen, P; Sneppen, Kim.
In: Physical Review Letters, Vol. 94, No. 2, 028701, 2005. -
In: Europhysics Letters, Vol. 69, No. 5, 03.2005, p. 853-859.
-
Functional alignment of regulatory networks: a study of temperate phages. / Trusina, Ala; Sneppen, Kim; Dodd, Ian B.; Shearwin, K. E.; Egan, J. B.In: PLoS Computational Biology, Vol. 1, No. 7, 2006.
Nuværende forskningsbevillinger:
-
StemPhys: Danish National Research Foundation Center for Stem Cell Decision Making (StemPhys)
60.000.000 Dkr granted by Danish National Research Foundation, 2015-2021.
Main PI: Lene B. Oddershede, Co-PIs Mogens H. Jensen, Ala Trusina, Elke Ober, Anne Grapin Botton, Joshua Brickman.
Ala Trusina, Associate Professor
Niels Bohr Building
Jagtvej 132, 2200 København N.
Email: trusina'at'nbi.ku.dk
Phone: +45 21 28 86 47
News on Biocomplexity and Biophysics
Staff
| Name | Title | Phone | |
|---|---|---|---|
| Search in Name | Search in Title | Search in Phone | |
| Trusina, Ala | Associate Professor | +4521288647 |
Group leader
Ala Trusina, Associate Professor
Email: trusina'at'nbi.ku.dk
Phone: +45 21 28 86 47
External staff & students
| Navn | Titel | Telefon | |
|---|---|---|---|
| Julie Klepstad | Master student | +45 | |
| Thomas Langemark Trojahn | Master student |